Abstract
Purpose
In the present work, we investigated the expression pattern of miR-4463 in the non-metastasis and metastasis colorectal cancer (CRC) patients and its regulation axis.
Methods
RT-qPCR assay was performed to assess miR-4463 expression in the serum and tissues of patients with non-metastasis and metastasis, and in the CRC cell lines. MTT assay, colony formation assay, transwell assay, and flow cytometry assay were used to examine the role of miR-4463 in CRC cell viability, proliferation, and migration. Bioinformatic analysis was used to identify the potential target gene of miR-4463, and the targeting relationship between miR-4463 and PPP1R12B was verified in vitro using dual luciferase assay. Western blotting assay was used to determine the protein level of the target gene PPP1R12B in CRC cells under the transfections of miR-4463 mimic, inhibitor and vectors overexpressing PPP1R12B.
Results
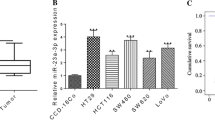
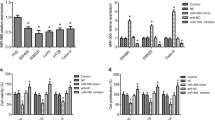
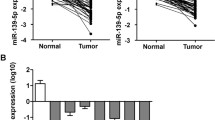
miR-4463 was markedly increased in the non-metastasis CRC tissues, and increased even higher in the metastasis CRC tissues, while miR-4463 expression had no significant difference in serum from non-metastasis and metastasis CRC samples. Besides, miR-4463 was upregulated in CRC cell lines. Functionally, miR-4463 promoted CRC cell proliferation, migration, and inhibiting cell apoptosis. Further analysis revealed that the miR-4463/PPP1R12B axis was responsible for the role of this miRNA.
Conclusion
We reported the roles of miR-4463 in CRC proliferation and migration, supporting that miR-4463 could be a potential predictive diagnostic marker for colon cancer.




Similar content being viewed by others
References
Marmol I, Sanchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18:197.
Cappell MS. From colonic polyps to colon cancer: pathophysiology, clinical presentation, and diagnosis. Clin Lab Med. 2005;25:135–77.
Cappell MS. From colonic polyps to colon cancer: pathophysiology, clinical presentation, screening and colonoscopic therapy. Minerva Gastroenterol Dietol. 2007;53:351–73.
Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–64.
Latham A, Srinivasan P, Kemel Y, Shia J, Bandlamudi C, Mandelker D, Middha S, Hechtman J, Zehir A, Dubard-Gault M, et al. microsatellite instability is associated with the presence of lynch syndrome pan-cancer. J Clin Oncol. 2019;37(4):286–95.
Akkoca AN, Yanık S, Özdemir ZT, Cihan FG, Sayar S, Cincin TG, Çam A, Özer C. TNM and Modified Dukes staging along with the demographic characteristics of patients with colorectal carcinoma. Int J Clin Exp Med. 2014;7:2828.
Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59:366–78.
Pinsky PF, Schoen RE. Contribution of surveillance colonoscopy to colorectal cancer prevention. Clin Gastroenterol Hepatol. 2020;18:2937-2944.e2931.
Gomes CP, Cho JH, Hood L, Franco OL, Pereira RW, Wang K. A review of computational tools in microRNA discovery. Front Genet. 2013;4:81.
Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer–a brief overview. Adv Biol Regul. 2015;57:1–9.
Hata A, Lieberman J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci Signal. 2015;8:re3.
Guo QR, Wang H, Yan YD, Liu Y, Su CY, Chen HB, Yan YY, Adhikari R, Wu Q, Zhang JY. The role of exosomal microRNA in cancer drug resistance. Front Oncol. 2020;10:472.
Zhang Y, Li M, Ding Y, Fan Z, Zhang J, Zhang H, Jiang B, Zhu Y. Serum MicroRNA profile in patients with colon adenomas or cancer. BMC Med Genomics. 2017;10:23.
Ding B, Yao M, Fan W, Lou W. Whole-transcriptome analysis reveals a potential hsa_circ_0001955/hsa_circ_0000977-mediated miRNA-mRNA regulatory sub-network in colorectal cancer. Aging (Albany NY). 2020;12:5259–79.
Wang J, Du Y, Liu X, Cho WC, Yang Y. MicroRNAs as regulator of signaling networks in metastatic colon cancer. Biomed Res Int. 2015;2015:823620.
Jaca A, Govender P, Locketz M, Naidoo R. The role of miRNA-21 and epithelial mesenchymal transition (EMT) process in colorectal cancer. J Clin Pathol. 2017;70:331–56.
Danac JMC, Uy AGG, Garcia RL. Exosomal microRNAs in colorectal cancer: Overcoming barriers of the metastatic cascade. Int J Mol Med. 2021;47:1–16.
Wei L, Chen Z, Cheng N, Li X, Chen J, Wu D, Dong M, Wu X. MicroRNA-126 inhibit viability of colorectal cancer cell by repressing mTOR induced apoptosis and autophagy. Onco Targets Ther. 2020;13:2459–68.
Hong YG, Xin C, Zheng H, Huang ZP, Yang Y, Zhou JD, Gao XH, Hao L, Liu QZ, Zhang W, et al. miR-365a-3p regulates ADAM10-JAK-STAT signaling to suppress the growth and metastasis of colorectal cancer cells. J Cancer. 2020;11:3634–44.
Huang L, Liu Z, Hu J, Luo Z, Zhang C, Wang L, Wang Z. MiR-377–3p suppresses colorectal cancer through negative regulation on Wnt/beta-catenin signaling by targeting XIAP and ZEB2. Pharmacol Res. 2020;156:104774.
Zhang Z, Li J, Huang Y, Peng W, Qian W, Gu J, Wang Q, Hu T, Ji D, Ji B, et al. Upregulated miR-1258 regulates cell cycle and inhibits cell proliferation by directly targeting E2F8 in CRC. Cell Prolif. 2018;51:e12505.
Li Y, Chen Z. The oncogenic role of microRNA-500a in colorectal cancer. Oncol Lett. 2020;19:1799–805.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Ahmed M. Colon cancer: a clinician’s perspective in 2019. Gastroenterol Res. 2020;13:1–10.
Burt RW, Barthel JS, Dunn KB, David DS, Drelichman E, Ford JM, Giardiello FM, Gruber SB, Halverson AL, Hamilton SR, et al. NCCN clinical practice guidelines in oncology. Colorectal cancer screening. J Natl Compr Canc Netw. 2010;8:8–61.
Eyking A, Reis H, Frank M, Gerken G, Schmid KW, Cario E. MiR-205 and MiR-373 are associated with aggressive human mucinous colorectal cancer. PLoS ONE. 2016;11:e0156871.
Zhou Y, Zhou G, Tian C, Jiang W, Jin L, Zhang C, Chen X. Exosome-mediated small RNA delivery for gene therapy. Wiley Interdiscip Rev RNA. 2016;7:758–71.
Balacescu O, Sur D, Cainap C, Visan S, Cruceriu D, Manzat-Saplacan R, Muresan MS, Balacescu L, Lisencu C, Irimie A. The impact of miRNA in colorectal cancer progression and its liver metastases. Int J Mol Sci. 2018;19:3711.
Li J, Peng W, Yang P, Chen R, Gu Q, Qian W, Ji D, Wang Q, Zhang Z, Tang J, et al. MicroRNA-1224-5p inhibits metastasis and epithelial-mesenchymal transition in colorectal cancer by targeting SP1-Mediated NF-kappaB signaling pathways. Front Oncol. 2020;10:294.
Wang X, Li H, Zhang Y, Liu Q, Sun X, He X, Yang Q, Yuan P, Zhou X. Suppression of miR-4463 promotes phenotypic switching in VSMCs treated with Ox-LDL. Cell Tissue Res. 2021;383:1155–65.
Hu T, Li J, Zhang C, Li S, He S, Yan H, Tan Y, Lei M, Wen M, Zuo J. The potential value of microRNA-4463 in the prognosis evaluation in hepatocellular carcinoma. Genes Dis. 2017;4:116–22.
Wang X, He X, Deng X, He Y, Zhou X. Roles of miR4463 in H2O2induced oxidative stress in human umbilical vein endothelial cells. Mol Med Rep. 2017;16:3242–52.
Morin PJ. Colorectal cancer: the APC-lncRNA link. J Clin Invest. 2019;129:503–5.
Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: designed to create specificity. Trends Biochem Sci. 2010;35:450–8.
Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33:537–45.
Okamoto R, Kato T, Mizoguchi A, Takahashi N, Nakakuki T, Mizutani H, Isaka N, Imanaka-Yoshida K, Kaibuchi K, Lu Z. Characterization and function of MYPT2, a target subunit of myosin phosphatase in heart. Cell Signal. 2006;18:1408–16.
Ding C, Tang W, Wu H, Fan X, Luo J, Feng J, Wen K, Wu G. The PEAK1–PPP1R12B axis inhibits tumor growth and metastasis by regulating Grb2/PI3K/Akt signalling in colorectal cancer. Cancer Lett. 2019;442:383–95.
Acknowledgements
We would like to thank all the researchers and study participants for their contributions.
Funding
This study was supported by the mechanism and expression of miR-4463 in patients with colorectal cancer (Project label: GCPY201710); A detection kit of miRNA for early diagnosis of colorectal cancer (Project label: ZHZD201801); Zhu Yong (QRX17092); Academic leader and talent cultivation (Project label: YRC2014-ZY3).
Author information
Authors and Affiliations
Contributions
JT, BJ, YZ and TL conceived the study. JT, TL and JX wrote the manuscript and designed the figures. YH, ZC, KZ and YD contributed to the writing and editing of the manuscript. All authors and participants reviewed the paper and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by Ethical Committee of The Nanjing Hospital of Chinese Medicine Affiliated to Nanjing University of Chinese Medicine and conducted in accordance with the ethical standards.
Informed consent
Yes.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

Rights and permissions
About this article
Cite this article
Tan, J., Lu, T., Xu, J. et al. MicroRNA-4463 facilitates the development of colon cancer by suppression of the expression of PPP1R12B. Clin Transl Oncol 24, 1115–1123 (2022). https://doi.org/10.1007/s12094-021-02752-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-021-02752-0