Abstract
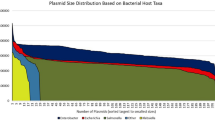
Plasmids play a fundamental role in the evolution of bacteria by allowing them to adapt to different environments and acquire, through horizontal transfer, genes that confer resistance to different classes of antibiotics. Using the available in vitro and in silico plasmid typing systems, we analyzed a set of isolates and public genomes of K. variicola to study its plasmid diversity. The resistome, the plasmid multilocus sequence typing (pMLST), and molecular epidemiology using the MLST system were also studied. A high frequency of IncF plasmids from human isolates but lower frequency from plant isolates were found in our strain collection. In silico detection revealed 297 incompatibility (Inc) groups, but the IncFIBK (216/297) predominated in plasmids from human and environmental samples, followed by IncFIIK (89/297) and IncFIA/FIA(HI1) (75/297). These Inc groups were associated with clinically important ESBL (CTX-M-15), carbapenemases (KPC-2 and NDM-1), and colistin-resistant genes which were associated with major sequence types (ST): ST60, ST20, and ST10. In silico MOB typing showed 76% (311/404) of the genomes contained one or more of the six relaxase families with MOBF being most abundant. We identified untypeable plasmids carrying blaKPC-2, blaIMP-1, and blaSHV-187 but for which a relaxase was found; this may suggest that novel plasmid structures could be emerging in this bacterial species. The plasmid content in K. variicola has limited diversity, predominantly composed of IncFIBK plasmids dispersed in different STs. Plasmid detection using the replicon and MOB typing scheme provide a broader context of the plasmids in K. variicola. This study showed that whole-sequence-based typing provides current insights of the prevalence of plasmid types and their association with antimicrobial resistant genes in K. variicola obtained from humans and environmental niches.





Similar content being viewed by others
Data availability
The draft genome sequences of K. variicola 06–268, 4880, 9635, and VI were deposited in GenBank under accession numbers; NZ_CXOZ00000000.1, CXPB00000000.1, and JAGMVR000000000.1, respectively.
References
Barrios-Camacho H, Aguilar-Vera A, Beltran-Rojel M et al (2019) Molecular epidemiology of Klebsiella variicola obtained from different sources. Sci Rep 9(1):10610. https://doi.org/10.1038/s41598-019-46998-9
Bi D, Zheng J, Li JJ, Sheng ZK, Zhu X, Ou HY, Li Q, Wei Q (2018) In silico typing and comparative genomic analysis of IncFIIK plasmids and insights into the evolution of replicons, plasmid backbones, and resistance determinant profiles. Antimicrob Agents Chemother 62(10):e00764-18. https://doi.org/10.1128/AAC.00764-18
Carattoli A (2009) Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53(6):2227–2238. https://doi.org/10.1128/AAC.01707-08
Carattoli A, Bertini A, Villa L et al (2005) Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63(3):219–228. https://doi.org/10.1016/j.mimet.2005.03.018
Carattoli A, Zankari E, García-Fernández A et al (2014) In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58(7):3895–3903. https://doi.org/10.1128/AAC.02412-14
Catalán-Nájera JC, Barrios-Camacho H, Duran-Bedolla J et al (2019) Molecular characterization and pathogenicity determination of hypervirulent Klebsiella pneumoniae clinical isolates serotype K2 in Mexico. Diagn Microbiol Infect Dis 94(3):316–319. https://doi.org/10.1016/j.diagmicrobio.2019.01.013. (Erratum in: Diagn Microbiol Infect Dis. 2020;96(1):114917)
Dong N, Yang X, Chan EW, Zhang R, Chen S (2022) Klebsiella species: taxonomy, hypervirulence and multidrug resistance. EBioMedicine 79:103998. https://doi.org/10.1016/j.ebiom.2022.103998
Duran-Bedolla J, Garza-Ramos U, Rodríguez-Medina N et al (2021) Exploring the environmental traits and applications of Klebsiella variicola. Braz J Microbiol 52(4):2233–2245. https://doi.org/10.1007/s42770-021-00630-z
Francisco A, Vaz C, Monteiro P et al (2012) PHYLOViZ: Phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinform 13:87. https://doi.org/10.1186/1471-2105-13-87
Garcillán-Barcia MP et al (2009) MP, Francia MV, de la Cruz F (2009) The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev 33(3):657–687. https://doi.org/10.1111/j.1574-6976.2009.00168.x
Garcillán-Barcia MP, Redondo-Salvo S, Vielva L et al (2020) MOBscan: automated annotation of MOB relaxases. Methods Mol Biol 2075:295–308. https://doi.org/10.1007/978-1-4939-9877-7_21
Garza-Ramos U, Silva-Sánchez J, Martínez-Romero E et al (2015) Development of a multiplex-PCR probe system for the proper identification of Klebsiella variicola. BMC Microbiol 15:64. https://doi.org/10.1186/s12866-015-0396-6
Garza-Ramos U, Rodriguez-Medina N, Lozano-Aguirre L et al (2021) Klebsiella variicola reference strain F2R9 (ATCC BAA-830) genome sequence. Microbiol Resour Announc 10(26):e0032921. https://doi.org/10.1128/MRA.00329-21
Gorrie CL, Mirceta M, Wick RR et al (2018) Antimicrobial-resistant Klebsiella pneumoniae carriage and infection in specialized geriatric care wards linked to acquisition in the referring hospital. Clin Infect Dis 67(2):161–170. https://doi.org/10.1093/cid/ciy027
Ito R, Mustapha MM, Tomich AD et al (2017) Widespread fosfomycin resistance in gram-negative bacteria attributable to the chromosomal fosA gene. mBio 8(4):e00749–17. https://doi.org/10.1128/mBio.00749-17.
Jiang Y, Yu D, Wei Z et al (2010) Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob Agents Chemother 54:3967–3969. https://doi.org/10.1128/AAC.00137-10
Long SW, Linson SE et al (2017) Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere 2(4):e00290–17. https://doi.org/10.1128/AAC.00175-10
Marasini D, Fakhr MK (2014) Exploring PFGE for detecting large plasmids in Campylobacter jejuni and Campylobacter coli isolated from various retail meats. Pathogens 3(4):833–844. https://doi.org/10.3390/pathogens3040833
Martínez-Romero E, Silva-Sanchez J, Barrios H et al (2015) Draft genome sequences of Klebsiella variicola plant isolates. Genome Announc 3(5):e01015-15. https://doi.org/10.1128/genomeA.01015-15
Mathers AJ, Peirano G, Pitout JDD (2015) The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrugresistant Enterobacteriaceae. Clin Microbiol Rev. https://doi.org/10.1128/CMR.00116-14
Osborn AM, da Silva Tatley FM, Steyn LM et al (2000) Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology 146(Part 9):2267–2275. https://doi.org/10.1099/00221287-146-9-2267
Potter RF, Lainhart W, Twentyman J et al (2018). Population structure, antibiotic resistance, and uropathogenicity of Klebsiella variicola. mBio 9:e02481–18. https://doi.org/10.1128/mBio.02481-18.
Rodríguez-Martínez JM, Díaz de Alba P, Briales A et al (2013) Contribution of OqxAB efflux pumps to quinolone resistance in extended-spectrum-β-lactamase-producing Klebsiella pneumoniae. J Antimicrob Chemother 68(1):68–73. https://doi.org/10.1093/jac/dks377
Rodríguez-Medina N, Barrios-Camacho H, Duran-Bedolla J et al (2019) Klebsiella variicola: an emerging pathogen in humans. Emerg Microbes Infect 8(1):973–988. https://doi.org/10.1080/22221751.2019.1634981
Rodríguez-Medina N, Martínez-Romero E, De la Cruz MA et al (2020) A Klebsiella variicola plasmid confers hypermucoviscosity-like phenotype and alters capsule production and virulence. Front Microbiol 11:579612. https://doi.org/10.3389/fmicb.2020.579612.
Rodríguez-Santiago J, Cornejo-Juárez P, Silva-Sánchez J et al (2021) Polymyxin resistance in Enterobacterales: overview and epidemiology in the Americas. Int J Antimicrob Agents 58(5):106426. https://doi.org/10.1016/j.ijantimicag.2021.106426
Rosenblueth M, Martinez L, Silva J, Martinez-Romero E (2004) Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst Appl Microbiol 27:27–35
Shankar C, MuthuirulandiSethuvel DP, Neeravi AR et al (2020) Identification of plasmids by PCR based replicon typing in bacteremic Klebsiella pneumoniae. Microb Pathog 148:104429. https://doi.org/10.1016/j.micpath.2020.104429
Smalla K, Jechalke S, Top EM (2015) Plasmid detection, characterization, and ecology. Microbiol Spectr 3(1):PLAS-0038–2014. https://doi.org/10.1128/microbiolspec.PLAS-0038-2014.
Tümmler B (2020) Molecular epidemiology in current times. Environ Microbiol 22(12):4909–4918. https://doi.org/10.1111/1462-2920.15238
Villa L, García-Fernández A, Fortini D et al (2010) Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65(12):2518–2529. https://doi.org/10.1093/jac/dkq347
Wang X, Zhao J, Ji F et al (2021) Multiple-replicon resistance plasmids of Klebsiella mediate extensive dissemination of antimicrobial genes. Front Microbiol 12:754931. https://doi.org/10.3389/fmicb.2021.754931
Wyres KL, Lam MMC, Holt KE (2020) Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol 18(6):344–359. https://doi.org/10.1038/s41579-019-0315-1
Yang X, Wai-Chi Chan E, Zhang R et al (2019) A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat Microbiol 4(12):2039–2043. https://doi.org/10.1038/s41564-019-0566-7
Acknowledgements
The authors thank Alejandro Sánchez Pérez for his technical assistance.
Funding
This work was supported by grant 347316 FORDECYT-PRONACES from SEP-CONACyT (Secretaría de Educación Pública-Consejo Nacional de Ciencia y Tecnología).
Author information
Authors and Affiliations
Contributions
Conceived and designed the study: JDB, NRM, and UGR. Performed the experiments: JDB, DMG, HBC, RSR, JART, and AAV. Analyzed and interpreted the data: JDB, NRM, MD, HBC, AAV, and UGR. Contributed reagents, materials, analysis tools, or data: RSR, EAV, JART, and UGR. Wrote the paper: JDB, NRM, and UGR. Revised and edited the paper: MD, NRM, and UGR.
Corresponding author
Ethics declarations
Research and ethics statement
The laboratory procedures carried out in this project with plant tissues were reviewed and approved by the research, biosafety, and ethics commissions of the National Institute of Public Health.
This project was exempt from review by the Ethic Commission at National Institute of Public Health because it does not involve human subjects and/or it is not an academic study and/or it does not include the analysis of data previously obtained from another study requiring the patients’ informed consent. On the other hand, the bacteria included in the study were obtained by routine procedures in each of the hospitals involved.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Josefina Duran-Bedolla and Nadia Rodríguez-Medina have contributed equally to this research.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Duran-Bedolla, J., Rodríguez-Medina, N., Dunn, M. et al. Plasmids of the incompatibility group FIBK occur in Klebsiella variicola from diverse ecological niches. Int Microbiol 26, 917–927 (2023). https://doi.org/10.1007/s10123-023-00346-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-023-00346-0