Abstract
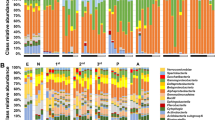
Soil inhabiting organisms are important determinants of agroecosystem productivity. Understanding the composition, the abundance, and the type of interactions established by soil microorganisms is therefore crucial to design strategies to improve agricultural practices and agroecosystem management. In this study, we collected Zeldia punctata nematodes in maize fields in South Africa and profiled their associated bacterial communities using next-generation sequencing. We observed that Z. punctata nematodes establish associations with ecologically diverse bacterial species. The most abundant species observed are Pseudomonas syringae, a phytopathogenic bacterial complex; Lactobacillus paraplantarum, a broadly distributed bacterial species that is present in soils, water bodies, and animal intestinal tracts and has certain probiotic and antimicrobial properties; and Melissococcus plutonius, a serious pathogenic bacterial species that causes brood disease in honeybees. Our study contributes to a better understanding of the soil bacterial communities associated with nematodes in maize agricultural soils in South Africa and unravels the presence of diverse detrimental and beneficial nematode-associated bacteria.





Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files. New generation sequencing data were deposited in the NCBI under the following biosample accession numbers: SAMN20693819 and SAMN20694755-57.
References
Abolafia J, Peña-Santiago R (2003) Nematodes of the order Rhabditida from Andalucía Oriental, Spain. The Genera Nothacrobeles Allen & Noffsinger, 1971 and Zeldia Thorne, 1937. J Nematol 35(3): 233–243.
Amirzadi N, Shokoohi E, Abolafia J (2013) Description of nine species of the family Cephalobidae (Nematoda, Rhabditida) and morphometric analysis in the genus Acrobeles von Linstow, 1877. Acta Zool Bul 65(1):3–26
Arce CC, Machado RAR, Ribas NS, Cristaldo PF, Ataíde LM, Pallini Â, Carmo FM, Freitas LG, Lima E (2017) Nematode root herbivory in tomato increases leaf defenses and reduces leaf miner oviposition and performance. J Chem Ecol 43(2):120–128. https://doi.org/10.1007/s10886-016-0810-z
Baquiran JP, Thater B, Sedky S, De Ley P, Crowley D, Orwin PM (2013) Culture independent investigation of the microbiome associated with the nematode Acrobeloides maximus. PLoS ONE 8:e67425. https://doi.org/10.1371/journal.pone.0067425
Bhat AH, Chaubey AK, Shokoohi E, Machado RAR (2020) Molecular and phenotypic characterization of Heterorhabditis indica (Nematoda: Rhabditida) nematodes isolated during a survey of agricultural soils in Western Uttar Pradesh, India. Acta Parasitol 1-17.https://doi.org/10.1007/s11686-020-00279-y
Blanc C, Sy M, Djigal D, Brauman A, Normand P, Villenave C (2006) Nutrition on bacteria by bacterial-feeding nematodes and consequences on the structure of soil bacterial community. Eur J Soil Biol 42:S70–S78
Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Caporaso JG (2018b) Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6(1):90. https://doi.org/10.1186/s40168-018-0470-z
Bokulich NA, Dillon MR, Zhang Y, Rideout JR, Bolyen E, Li H, Albert PS, Caporaso JG (2018) q2-longitudinal: longitudinal and paired-sample analyses of microbiome data. mSystems 3(6): e00219–18. https://doi.org/10.1128/mSystems.00219-18.
Bolyen E, Rideout JR, Dillonm MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37(8):852–857. https://doi.org/10.1038/s41587-019-0209-9
Budge G, Shirley M, Jones B, Quill E, Tomkies V, Feil EJ, Brown MA, Haynes EG (2014) Molecular epidemiology and population structure of the honey bee brood pathogen Melissococcus plutonius. ISME J 8:1588–1597. https://doi.org/10.1038/ismej.2014.20
Bull CT, De Boer SH, Denny TP, Firrao G, Fischer-Le Saux M, Saddler GS, Scortichini M, Stead DE, Takikawa Y (2010) Comprehensive list of names of plant pathogenic bacteria, 1980–2007. J Plant Pathol 92:551–592
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13(7):581–583. https://doi.org/10.1038/nmeth.3869
Chevenet F, Brun C, Banuls AL, Jacq B, Christen R (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439
De Grisse A (1969) Redescription ou modifications de quelques techniques utilisées dans l’étude des nematodes phytoparasitaire]. Mededelingen Rijksfaculteit Der Landbouwwetenschappen Gent 34:351–369
De Ley P (1992) The nematode community of a marginal soil at Cambérène, Senegal, with special attention to functional morphology and niche partitioning in the family Cephalobidae. Mededelingen Van De Koninklijke Academie Voor Wetenschappen, Letteren En Schone Kunsten Van België - Klasse Der Wetenschappen 53:107–153
De Ley P, Felix MA, Frisse LM, Nadler SA, Sternberg PW, Thomas WK (1999) Molecular and morphological characterisation of two reproductively isolated species with mirror-image anatomy (Nematoda: Cephalobidae). Nematology 2:591–612. https://doi.org/10.1163/156854199508559
Dirksen P, Marsh SA, Braker I, Heitland N, Wagner S, Nakad R, Mader S, Petersen C, Kowallik V, Rosenstiel P, Félix MA, Schulenburg H (2016) The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol 14:38. https://doi.org/10.1186/s12915-016-0258-1
Djigal D, Sy M, Brauman A, Diop T, Mountport D, Chotte J, Villenave C (2004a) Interactions between Zeldia punctata (Cephalobidae) and bacteria in the presence or absence of maize plants. Plant Soil 262:33–44. https://doi.org/10.1023/B:PLSO.0000037022.16822.75
Djigal D, Brauman A, Diop TA, Chotte JL, Villenave C (2004b) Influence of bacterial-feeding nematodes (Cephalobidae) on soil microbial communities during maize growth. Soil Biol Biochem 36:323–331
Douglas AE (2009) The microbial dimension in insect nutritional ecology. Funct Ecol 23(1):38–47
Elhady A, Giné A, Topalovic O, Jacquiod S, Sørensen SJ, Sorribas FJ, Holger H (2017) Microbiomes associated with infective stages of root-knot and lesion nematodes in soil. PLoS ONE 12(5):e0177145. https://doi.org/10.1371/journal.pone.0177145
Entry JA, Mills DK, Mathee K, Jayachandran K, Sojka RE, Narasimhan G (2008) Influence of irrigated agriculture on soil microbial diversity. Appl Soil Ecol 40:146–154
Esser RP (1972) Effect of sodium hypochlorite concentrations on selected genera of nematodes. Proc Helmint Soc Wash 39(1):108–114
Faith DP (1992) Conservation evaluation and phylogenetic diversity. Biol Cons 61:1–10
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88(6):1354–1364
Gaugler R (2002) Entomopathogenic Nematology, CABI. 388 pp.
Glick BR, Cheng Z, Czarny J, Duan J (2007) Promotion of plant growth by ACC deaminase-producing soil bacteria. In: Bakker, P.A.H.M., Raaijmakers, J.M., Bloemberg, G., Höfte, M., Lemanceau, P., Cooke, B.M. (eds) New perspectives and approaches in plant growth-promoting rhizobacteria research. Springer, Dordrecht. https://doi.org/10.1007/978-1-4020-6776-1_8
Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579. https://doi.org/10.1038/ismej.2011.41
Kabouw P, Kos M, Kleine S, Vockenhuber EA, van Loon JJ, Van der Putten WH, Van Dam NM, Biere A (2011) Effects of soil organisms on aboveground multitrophic interactions are consistent between plant genotypes mediating the interaction. Entom Experi Et Applicata 139(3):197–206
Kaplan F, Badri DV, Zachariah C, Ajredini R, Sandoval FJ, Roje S, Levine LH, Zhang F, Robinette SL, Alborn HT, Zhao W, Stadler M, Nimalendran R, Dossey AT, Brüschweiler R, Vivanco JM, Edison AS (2009) Bacterial attraction and quorum sensing inhibition in Caenorhabditis elegans exudates. J Chem Ecol 35(8):878–892. https://doi.org/10.1007/s10886-009-9670-0
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30(14):3059–3066. https://doi.org/10.1093/nar/gkf436
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33 (7): 1870–1874.
Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 8: 44 (W1), W242–5. https://doi.org/10.1093/nar/gkw290.
Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73(5):1576–1585. https://doi.org/10.1128/AEM.01996-06
Machado RAR, Arce CCM, McClure MA, Baldwin IT, Erb M (2018) Aboveground herbivory induced jasmonates disproportionately reduce plant reproductive potential by facilitating root nematode infestation. Plant Cell Environ 41(4):797–808. https://doi.org/10.1111/pce.13143
Machado RAR, Muller A, Ghazal S, Thanwisai A, Pages S, Bode HB, Hussein MA, Khalil KM, Tisa LS (2021) Photorhabdus heterorhabditis subsp. aluminescens subsp. nov., Photorhabdus heterorhabditis subsp. heterorhabditis subsp. nov., Photorhabdus australis subsp. thailandensis subsp. nov., Photorhabdus australis subsp. australis subsp. nov., and Photorhabdus aegyptia sp. nov. isolated from Heterorhabditis entomopathogenic nematodes. Inter J Sys Evol Microbiol 71(1): ff. https://doi.org/10.1099/ijsem.0.004610ff.ffhal-03143425f
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. https://doi.org/10.1038/ismej.2011.139
Meidong R, Nakao M, Sakai K, Tongpim S (2021) Lactobacillus paraplantarum L34b–2 derived from fermented food improves the growth, disease resistance and innate immunity in Pangasius bocourti. Aquaculture 531:735878. https://doi.org/10.1016/j.aquaculture.2020.735878
Mihailović M, Živković M, Jovanović JA, Tolinački M, Sinadinović M, Rajić J, Uskoković A, Dinić S, Grdović N, Golić N, Vidaković M (2017) Oral administration of probiotic Lactobacillus paraplantarum BGCG11 attenuates diabetes-induced liver and kidney damage in rats. J Func Foods 38:427–437. https://doi.org/10.1016/j.jff.2017.09.033
Nannipieri P, Ascher J, Ceccherini MT, Landi L, Pietramellara G, Renella G (2003) Microbial diversity and soil functions. European J Soil Sci 54:655–670. https://doi.org/10.1046/j.1351-0754.2003.0556.x
Neal AL, Ahmad S, Gordon-Weeks R, Ton J (2012) Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS ONE 7(4):e35498. https://doi.org/10.1371/journal.pone.0035498
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, Oxford
Ogier JC, Pagès S, Frayssinet M, Gaudriault S (2020) Entomopathogenic nematode-associated microbiota: from monoxenic paradigm to pathobiome. Microbiome 8(1):25. https://doi.org/10.1186/s40168-020-00800-5
Parr JF, Papendick RI, Hornick SB, Meyer RE (1992) Soil quality: attributes and relationship to alternative and sustainable agriculture. American J Alte Agric 7:5–11
Price MN, Dehal PS, Arkin AP (2010) FastTree 2–approximately maximum‐likelihood trees for large alignments. PLoS ONE 5:e9490.
Rudrappa T, Czymmek KJ, Paré PW, Bais HP (2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148(3):1547–1556. https://doi.org/10.1104/pp.108.127613
Shokoohi E, Abolafia J (2019) Soil and freshwater rhabditid nematodes (Nematoda, Rhabditida) from Iran: A compendium. Universidad de Jaén, Spain, p 224
Shokoohi E, Abolafia J, Mashela PW (2020) Redescription of Paratrophurus anomalus from South Africa. Nematology 22(5):543–554. https://doi.org/10.1163/15685411-00003322
Shokoohi E (2021) Morphological and molecular characters of Scutellonema brachyurus (Steiner, 1938) Andrassy, 1958 from South Africa. J Nematol 53: 1–13. https://doi.org/10.21307/jofnem-2021-027
Straube D, Juen A (2013) Storage and shipping of tissue samples for DNA analyses: a case study on earthworms. European J Soil Biol 57:13–18
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol 9:678–687
Treonis AM, Michelle EH, O’Leary CA, Austin EE, Marks CB (2010) Identification and localization of food-source microbial nucleic acids inside soil nematodes. Soil Biol Biochem 42(11):2005–2011. https://doi.org/10.1016/j.soilbio.2010.07.026
Tulini FL, Winkelströter LK, De Martinis EC (2013) Identification and evaluation of the probiotic potential of Lactobacillus paraplantarum FT259, a bacteriocinogenic strain isolated from Brazilian semi-hard artisanal cheese. Anaerobe 22:57–63. https://doi.org/10.1016/j.anaerobe.2013.06.006
Villenave C, Bongers T, Ekschmitt K, Djigal D, Chotte JL (2001) Changes in nematode communities following cultivation of soils after fallow periods of different length. Appl Soil Ecol 17:43–52
Wagg C, Bender SF, Widmer F, van der Heijden MGA (2014) Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc National Acad Sci 111(14):5266–5270. https://doi.org/10.1073/pnas.1320054111
Winkelströter LK, Tulini FL, De Martinis ECP (2015) Identification of the bacteriocin produced by cheese isolate Lactobacillus paraplantarum FT259 and its potential influence on Listeria monocytogenes biofilm formation. LWT-Food Sci Technol 64(2):586–592
Xin XF, Kvitko B, He SY (2018) Pseudomonas syringae: what it takes to be a pathogen. Nat Rev Microbiol 16(5):316–328. https://doi.org/10.1038/nrmicro.2018.17
Yeates GW (2003) Nematodes as soil indicators: functional and biodiversity aspects. Biol Fertil Soils 37(4):199–210
Zhang F, Berg M, Dierking K, Félix MA, Shapira M, Samuel BS, Schulenburg H (2017) Caenorhabditis elegans as a model for microbiome research. Front Microbiol 23(8):485. https://doi.org/10.3389/fmicb.2017.00485
Acknowledgements
The authors thank the Technology Innovation Agency of the Republic of South Africa, the Institute of Biology of the University of Neuchatel, and the Swiss National Science Foundation for supporting the project.
Funding
This research was funded by the Technology Innovation Agency (TIA SEED FUND) of the Republic of South Africa. The work of RARM is supported by the Swiss National Science Foundation (Grant No. 186094 to RARM).
Author information
Authors and Affiliations
Contributions
Ebrahim Shokoohi conceptualized the project, designed experiments, analysed data, and wrote and revised the manuscript. PW Mashela edited and revised the manuscript. Ricardo A. R. Machado analysed data and wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.


10123_2021_207_MOESM2_ESM.jpg
Supplementary file2 Fig. S2 Alpha diversity for Z. punctata collected from maize fields using Shannon entropy. (JPG 30 KB)

10123_2021_207_MOESM3_ESM.jpg
Supplementary file3 Fig. S3 Relative frequency of the most abundant bacterial phyla associated with Z. punctata nematodes collected from maize fields in South Africa. (JPG 71 KB)

10123_2021_207_MOESM4_ESM.jpg
Supplementary file4 Fig. S4 Relative frequency of the most abundant bacterial orders associated with Z. punctata nematodes collected from maize fields in South Africa. (JPG 103 KB)

10123_2021_207_MOESM5_ESM.jpg
Supplementary file5 Fig. S5 Relative frequency of the most abundant bacterial families associated with Z. punctata nematodes collected from maize fields. (JPG 123 KB)
Rights and permissions
About this article
Cite this article
Shokoohi, E., Mashela, P.W. & Machado, R.A.R. Bacterial communities associated with Zeldia punctata, a bacterivorous soil-borne nematode. Int Microbiol 25, 207–216 (2022). https://doi.org/10.1007/s10123-021-00207-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-021-00207-8